By Jonathan MacQuitty & Galym Imanbayev
Amid the record-breaking fundraising activity in the therapeutics space, it is important to remember that 97% of cancer drugs that have entered clinical trials were never approved.¹ Among those that were, the median response rate to treatment continues to disappoint at 41%, with many of the latest successful immuno-oncology drugs having much lower response rates.² It’s a painful reality for patients who can suffer severe side effects just for the possibility of some therapeutic benefit, and for the healthcare system we all directly or indirectly pay into. These disappointing statistics reveal that we do not yet understand the biological complexity of cancer. We believe two critical requirements are needed to advance the field and lead to better, more precise patient treatment.
First, we must understand the deep and dynamic nature of cancer’s mutational heterogeneity and concurrently the patient’s immune response to the tumor. Second, we must develop novel approaches to predicting responses and thus guide precision treatment. These two foundational pillars of functional genomics and their importance have been at the core of several of our long-standing and more recent investments, an underlying ethos of which has been to skate to where the puck is going, not to where it is.
It is with that aspiration we elaborate further. The first challenge is largely a data problem. To date, most approaches have taken a static and narrow approach to data generation, building products to address where the technology and needs were 5–6 years ago, looking at just several hundred genes at any point in time. To fully understand cancer, we must move beyond rare data points and towards trend-lines and their first and second derivatives that may better elucidate and predict progression of cancer.
We believe no other company embodies this approach more than Personalis, a Lightspeed portfolio company that we’ve backed since the Series A. Since inception, Personalis has taken a rigorously comprehensive approach — building technology that enables the analysis of not only the whole exome covering over 20,000 genes, but also interrogates the transcriptome, T cell and B cell receptor repertoire profiling, and has potential to uncover novel neoantigen and biomarkers (Figure 1.). We believe the depth and breadth of this approach is the future of cancer analysis. These data enable biopharma companies and clinicians to better understand the mechanisms by which cancer develops and could be treated.
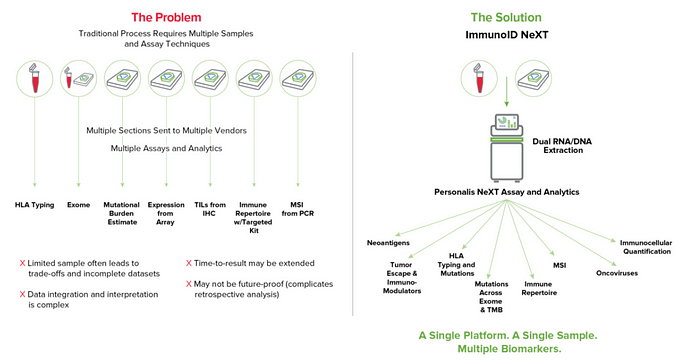
Personalis has also developed a proprietary composite biomarker that combines immune response and resistance mechanisms to better predict patient response to therapy. In addition, Personalis is able to track minimal residual disease through blood analysis. Their approach is 10–100x more sensitive than others and has a goal to detect possible cancer recurrence shortly after surgical resection. This can lead to detection over a year earlier, potentially enough to enable a second (conclusive) surgery or earlier drug treatment leading to dramatically improved clinical outcomes. This system also has the ability to measure cancer mutations that arise in response to the treatment which can inform clinical decisions to change a patient’s therapy when a tumor has begun to evade it. With nine of the top ten major biopharma companies as customers, more than 200,000 proprietary samples analyzed for several population sequencing initiatives, and multiple partnerships and collaborations with the likes of Mayo Clinic, we’re excited to support Personalis in raising the bar in genomics across both drug development and diagnostics.
Alongside the need for comprehensive human data, realizing the dream of precision oncology requires better functional genomics methods to uncover the vulnerabilities of cancer. The abysmal response rates to even the latest drugs is an unfortunate reminder that neither existing human clinical data nor cell line-based system are sufficient to predict treatment responses. Game changing innovation in functional genomics must genuinely recapitulate the biology of cancer in vivo (Figure 2), be high throughput, and be deeply quantifiable.
We believe that D2G Oncology, a Lightspeed portfolio company, has such a unique platform. D2G has developed a genetically engineered mouse model system that enables the multiplexed and quantitative analysis of cancer growth in vivo. Their systems enable the performance of various drugs and drug combinations to be evaluated across cancers of many genotypes in parallel. This allows identification of cancer genes that drive resistance or sensitivity to different drugs and is allowing D2G to build an expansive data platform. This data should lead to the more effective use of drugs and meaningfully de-risked clinical trials for new therapies and combination therapies. Ultimately, this approach should transform precision cancer therapeutics. We’re thrilled to support the team at D2G in spearheading this leap in functional cancer genomics.
While there is much work ahead, progress on these critical genomic capabilities will lead to massive improvements in patient outcomes. At Lightspeed, we strive to support patient-centric innovation rooted in scientific and technical breakthroughs and believe in teams who have a vision to transform patient outcomes at scale. With our investments in Personalis and D2G Oncology, we’re excited to be a part of building the foundation of the next chapter of functional genomics and to see the cancer world converging to where the puck was heading all along.
[1] “Estimation of clinical trial success rates and related parameters.” 31 Jan. 2018, https://academic.oup.com/biostatistics/article/20/2/273/4817524.
[2] “An Overview of Cancer Drugs Approved by the US Food and Drug ….” 28 May. 2019, https://jamanetwork.com/journals/jamainternalmedicine/fullarticle/2733563
[3] Winters IP, Murray CW, Winslow MM. Towards quantitative and multiplexed in vivo functional cancer genomics. Nat Rev Genet. 2018.
Authors



